Decoding the Genetic Interplay: Integrated Computational Insights into the Interplay Between Peri-Implantitis and Osteoporosis
Abstract
Background: Peri-implantitis (PI) and osteoporosis (OP) are prevalent chronic conditions with substantial health impacts, yet the genetic and molecular links between these diseases are not fully understood. Clarifying the shared genetic similarities between PI and OP is crucial for enhancing diagnostic and therapeutic strategies.
Objective: The study aims to elucidate the shared genetic and molecular pathways between PI and OP through an integrated computational biology analysis, contributing to the development of more effective diagnostic and therapeutic approaches.
Methods: The research utilized PI-related datasets GSE33774 and GSE106090 from the GEO database and genes associated with PI and OP from DisGeNET. Differential expression analysis was conducted using the "limma" package in R. Protein-Protein Interaction (PPI) networks were constructed with Cytoscape, identifying cross-talk genes between PI and OP. The LASSO model was applied for marker gene selection and GO Biological Process and KEGG pathway analyses were conducted. Analyses of drug susceptibility and immune infiltration were also included to understand potential therapeutic implications and the role of the immune environment.
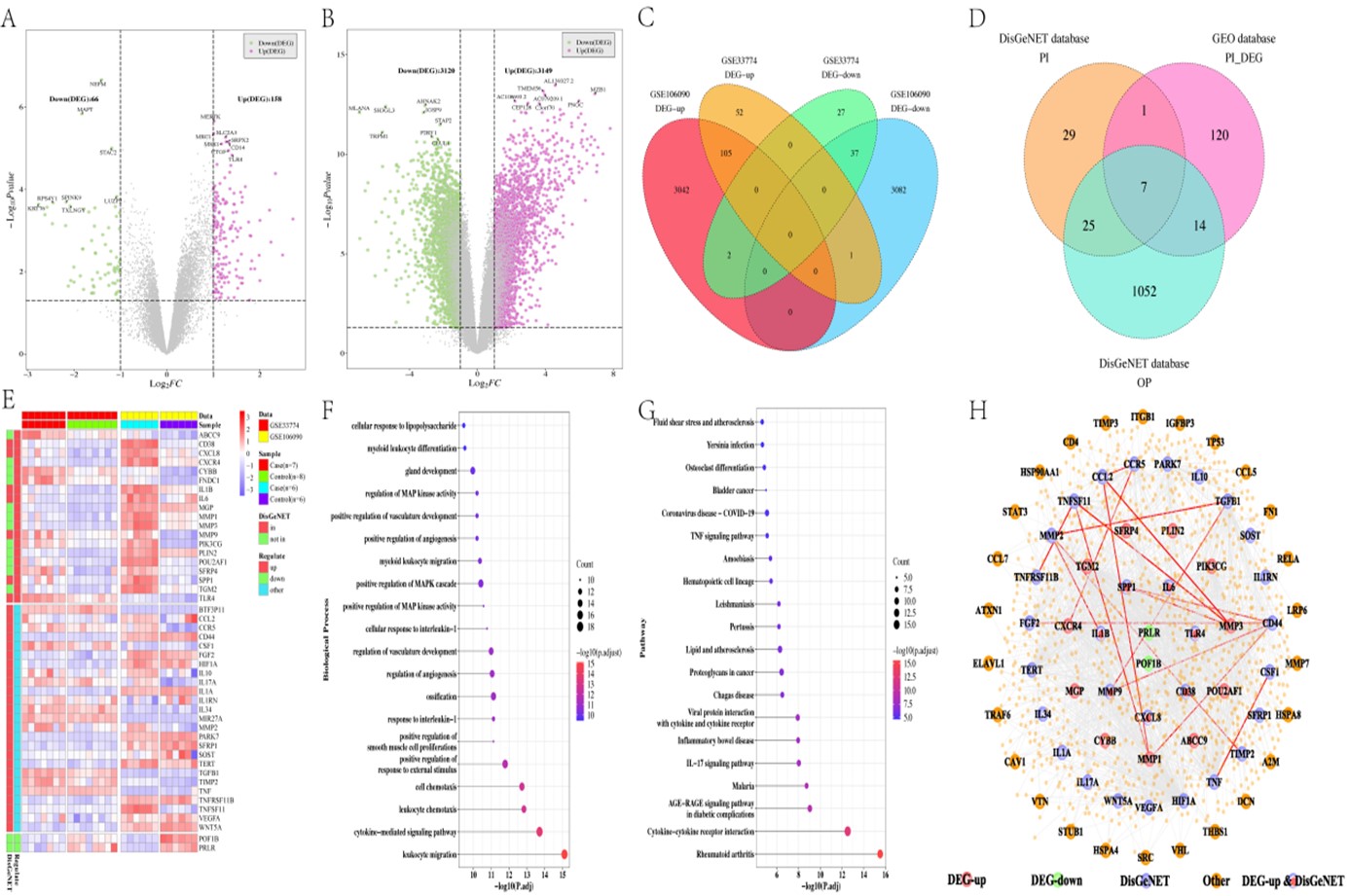
Results: The analysis identified key differentially expressed genes (DEGs) in PI and significant cross-talk genes between PI and OP. The PPI network highlighted central genes like HIF1A, TNF, TGM2, and SPP1. Marker genes (i.e., PIK3CG, SFRP4, CCR5, and PRLR) pinpointed through LASSO and logistic regression showed significant correlations with the cross-talk genes. Insights into drug susceptibility and the immune infiltration landscape in both conditions were provided. Single cell analysis further delineated the expression patterns of these marker genes in relevant cell types.
Conclusion: This study uncovers novel insights into the shared genetic landscape of peri-implantitis (PI) and osteoporosis (OP) by identifying four key marker genes—PIK3CG, SFRP4, CCR5, and PRLR—and their roles in the pathogenesis of these conditions. The integrated computational biology approach employed in this research contributes to the current body of literature by offering a deeper understanding of the genetic and molecular mechanisms underlying the interplay between PI and OP.
References
[2] De Medeiros, F., Kudo, G. A. H., Leme, B. G., Saraiva, P. P., Verri, F. R., Honório, H. M., et al. (2018). Dental implants in patients with osteoporosis: a systematic review with meta-analysis. International Journal of Oral and Maxillofacial Surgery, 47(4), 480–491. https://doi.org/10.1016/j.ijom.2017.05.021
[3] Guobis, Z., Pacauskiene, I., & Astramskaite, I. (2016). General diseases influence on peri-implantitis development: a systematic review. Journal of Oral and Maxillofacial Research, 7(3). https://doi.org/10.5037/jomr.2016.7305
[4] Dvorak, G., Arnhart, C., Heuberer, S., Huber, C. D., Watzek, G., & Gruber, R. (2011). Peri-implantitis and late implant failures in postmenopausal women: a cross-sectional study: Peri-implantitis in postmenopausal women. Journal of Clinical Periodontology, 38(10), 950–955. https://doi.org/10.1111/j.1600-051X.2011.01772.x
[5] Dreyer, H., Grischke, J., Tiede, C., Eberhard, J., Schweitzer, A., Toikkanen, S. E., et al. (2018). Epidemiology and risk factors of peri‐implantitis: A systematic review. Journal of Periodontal Research, 53(5), 657–681. https://doi.org/10.1111/jre.12562
[6] Corcuera‐Flores, J. R., Alonso‐Domínguez, A. M., Serrera‐Figallo, M. Á., Torres‐Lagares, D., Castellanos‐Cosano, L., & Machuca‐Portillo, G. (2016). Relationship between osteoporosis and marginal bone loss in osseointegrated implants: A 2‐year retrospective study. Journal of Periodontology, 87(1), 14–20. https://doi.org/10.1902/jop.2015.150229
[7] Smeets, R., Henningsen, A., Jung, O., Heiland, M., Hammächer, C., & Stein, J. M. (2014). Definition, etiology, prevention and treatment of peri-implantitis – a review. Head & Face Medicine, 10(1), 34. https://doi.org/10.1186/1746-160X-10-34
[8] Schwarz, F., Derks, J., Monje, A., & Wang, H. (2018). Peri‐implantitis. Journal of Clinical Periodontology, 45(S20). https://doi.org/10.1111/jcpe.12954
[9] Clowes, J. A., Riggs, B. L., & Khosla, S. (2005). The role of the immune system in the pathophysiology of osteoporosis. Immunological Reviews, 208(1), 207–227. https://doi.org/10.1111/j.0105-2896.2005.00334.x
[10] Kensara, A., Hefni, E., Williams, M. A., Saito, H., Mongodin, E., & Masri, R. (2021). Microbiological profile and human immune response associated with peri‐implantitis: A systematic review. Journal of Prosthodontics, 30(3), 210–234. https://doi.org/10.1111/jopr.13270
[11] Pietschmann, P., Mechtcheriakova, D., Meshcheryakova, A., Föger-Samwald, U., & Ellinger, I. (2016). Immunology of osteoporosis: a mini-review. Gerontology, 62(2), 128–137. https://doi.org/10.1159/000431091
[12] Petković, A. B., Matić, S. M., Stamatović, N. V., Vojvodić, D. V., Todorović, T. M., Lazić, Z. R., et al. (2010). Proinflammatory cytokines (IL-1β and TNF-α) and chemokines (IL-8 and MIP-1α) as markers of peri-implant tissue condition. International Journal of Oral and Maxillofacial Surgery, 39(5), 478–485. https://doi.org/10.1016/j.ijom.2010.01.014
[13] Zheng, S. X., Vrindts, Y., Lopez, M., De Groote, D., Zangerlé, P. F., Collette, J., et al. (1997). Increase in cytokine production (IL-1β, IL-6, TNF-α but not IFN-γ, GM-CSF or LIF) by stimulated whole blood cells in postmenopausal osteoporosis. Maturitas, 26(1), 63–71. https://doi.org/10.1016/S0378-5122(96)01080-8
[14] Contaldo, M., Itro, A., Lajolo, C., Gioco, G., Inchingolo, F., & Serpico, R. (2020). Overview on osteoporosis, periodontitis and oral dysbiosis: the emerging role of oral microbiota. Applied Sciences, 10(17), 6000. https://doi.org/10.3390/app10176000
[15] Hernández‐Vigueras, S., Martínez‐Garriga, B., Sánchez, M. C., Sanz, M., Estrugo‐Devesa, A., Vinuesa, T., et al. (2016). Oral microbiota, periodontal status, and osteoporosis in postmenopausal females. Journal of Periodontology, 87(2), 124–133. https://doi.org/10.1902/jop.2015.150365
[16] Sgolastra, F., Petrucci, A., Severino, M., Gatto, R., & Monaco, A. (2015). Smoking and the risk of peri‐implantitis: A systematic review and meta‐analysis. Clinical Oral Implants Research, 26(4). https://doi.org/10.1111/clr.12319
[17] Ratajczak, A. E., Szymczak-Tomczak, A., Rychter, A. M., Zawada, A., Dobrowolska, A., & Krela-Kaźmierczak, I. (2021). Impact of cigarette smoking on the risk of osteoporosis in inflammatory bowel diseases. Journal of Clinical Medicine, 10(7), 1515. https://doi.org/10.3390/jcm10071515
[18] Li, S., Zhou, C., Pelekos, G., Ziebolz, D., Schmalz, G., & Qin, Z. (2021). Similarity and potential relation between periimplantitis and rheumatoid arthritis on transcriptomic level: Results of a bioinformatics study. Frontiers in Immunology, 12, 702661. https://doi.org/10.3389/fimmu.2021.702661
[19] Lei, H., Chen, X., Wang, Z., Xing, Z., Du, W., Bai, R., et al. (2023). Exploration of the underlying comorbidity mechanism in psoriasis and periodontitis: A bioinformatics analysis. Hereditas, 160(1), 7. https://doi.org/10.1186/s41065-023-00266-z
[20] He, Z., Jiang, Q., Li, F., & Chen, M. (2021). Crosstalk between venous thromboembolism and periodontal diseases: A bioinformatics analysis. Disease Markers, 2021. https://doi.org/10.1155/2021/1776567
[21] Wang, X., Shi, N., Wu, B., Yuan, L., Chen, J., Ye, C., et al. (2022). Bioinformatics analysis of gene expression profile and functional analysis in periodontitis and Parkinson’s disease. Frontiers in Aging Neuroscience, 14, 1029637. https://doi.org/10.3389/fnagi.2022.1029637
[22] Kang, J., Kwon, E. J., Yu, Y., Kim, Y., & Kim, Y. H. (2022). Identification of shared genes and pathways in periodontitis and type 2 diabetes by bioinformatics analysis. Frontiers in Endocrinology, 12, 724278. https://doi.org/10.3389/fendo.2021.724278
[23] Liu, S., Wang, Z., Zhu, R., Wang, F., Cheng, Y., & Liu, Y. (2021). Three differential expression analysis methods for RNA sequencing: limma, EdgeR, DESeq2. Journal of Visualized Experiments, (175), e62528. https://doi.org/10.3791/62528
[24] Kolde, R., & Kolde, M. R. (2015). Package ‘pheatmap’. R Package, 1(7), 790.
[25] Harrell Jr, F. E., Harrell Jr, M. F. E., & Hmisc, D. (2017). Package ‘rms’. Vanderbilt University, 229, Q8.
[26] Bi, G., Li, R., Liang, J., Hu, Z., & Zhan, C. (2020). A nomogram with enhanced function facilitated by nomogramEx and nomogramFormula. Annals of Translational Medicine, 8(4). https://doi.org/10.21037/atm.2020.01.71
[27] Luo, W., & Brouwer, C. (2013). Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics, 29(14), 1830–1831. https://doi.org/10.1093/bioinformatics/btt285
[28] Venza, I., Visalli, M., Cucinotta, M., De Grazia, G., Teti, D., & Venza, M. (2010). Proinflammatory gene expression at chronic periodontitis and peri‐implantitis sites in patients with or without type 2 diabetes. Journal of Periodontology, 81(1), 99–108. https://doi.org/10.1902/jop.2009.090358
[29] Farangis, F., Majid, M., Gholamhossein, H., Soudeh, K., Hossein, K., Zahra, A., et al. (2017). CC chemokines CCL2, CCL3, CCL4 and CCL5 are elevated in osteoporosis patients. Journal of Biomedical Research, 31(5), 468. https://doi.org/10.7555/JBR.31.20150166
[30] Venable, J. D., Ameriks, M. K., Blevitt, J. M., Thurmond, R. L., & Fung-Leung, W. P. (2010). Phosphoinositide 3-kinase gamma (PI3Kγ) inhibitors for the treatment of inflammation and autoimmune disease. Recent Patents on Inflammation & Allergy Drug Discovery, 4(1), 1–15. https://doi.org/10.2174/187221310789895603
[31] Yu, T., Acharya, A., Mattheos, N., Li, S., Ziebolz, D., Schmalz, G., et al. (2019). Molecular mechanisms linking peri-implantitis and type 2 diabetes mellitus revealed by transcriptomic analysis. PeerJ, 7, e7124. https://doi.org/10.7717/peerj.7124
[32] Chai, Y., Tan, F., Ye, S., Liu, F., & Fan, Q. (2019). Identification of core genes and prediction of miRNAs associated with osteoporosis using a bioinformatics approach. Oncol Lett, 17(1), 468–481. https://doi.org/10.3892/ol.2018.9508
[33] Chen, R., Baron, R., & Gori, F. (2022). Sfrp4 and the biology of cortical bone. Curr Osteoporos Rep, 20(2), 153–161. https://doi.org/10.1007/s11914-022-00727-w
[34] Brommage, R., & Ohlsson, C. (2018). Translational studies provide insights for the etiology and treatment of cortical bone osteoporosis. Best Pract Res Clin Endocrinol Metab, 32(3), 329–340. https://doi.org/10.1016/j.beem.2018.02.006
[35] Haraguchi, R., Kitazawa, R., Mori, K., Tachibana, R., Kiyonari, H., Imai, Y., et al. (2016). sFRP4-dependent Wnt signal modulation is critical for bone remodeling during postnatal development and age-related bone loss. Sci Rep, 6(1), 25198. https://doi.org/10.1038/srep25198
[36] Lee, D. Y., Kim, H., Ku, S. Y., Kim, S. H., Choi, Y. M., & Kim, J. G. (2010). Association between polymorphisms in Wnt signaling pathway genes and bone mineral density in postmenopausal Korean women. Menopause, 17(5), 1064–1070. https://doi.org/10.1097/gme.0b013e3181da4da3
[37] Liang, J., Deng, Y., Zhang, Y., Wu, B., & Zhou, J. (2022). PRLR and CACNA2D1 impact the prognosis of breast cancer by regulating tumor immunity. J Pers Med, 12(12), 2086. https://doi.org/10.3390/jpm12122086
[38] Suarez, A. L. P., López-Rincón, G., Martínez Neri, P. A., & Estrada-Chávez, C. (2015). Prolactin in inflammatory response. In M. Diakonova (Ed.), Recent advances in prolactin research (Vol. 846, pp. 243–264). Cham: Springer International Publishing. https://doi.org/10.1007/978-3-319-12114-7_11
[39] Zhou, H., Chen, D., Xie, G., Li, J., Tang, J., & Tang, L. (2020). lncRNA-mediated ceRNA network was identified as a crucial determinant of differential effects in periodontitis and periimplantitis by high-throughput sequencing. Clin Implant Dent Relat Res, 22(3), 424–450. https://doi.org/10.1111/cid.12911
[40] Oh, J., Kim, Y., Son, H., Kim, Y. H., & Kim, H. (2023). Comparative transcriptome analysis of periodontitis and peri-implantitis in human subjects. J Periodontol, Oct 3, JPER.23-0289.
[41] Glaviano, A., Foo, A. S. C., Lam, H. Y., Yap, K. C. H., Jacot, W., Jones, R. H., et al. (2023). PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer, 22(1), 138. https://doi.org/10.1186/s12943-023-01827-6
[42] Moon, J. B., Kim, J. H., Kim, K., Youn, B. U., Ko, A., Lee, S. Y., et al. (2012). Akt induces osteoclast differentiation through regulating the GSK3β/NFATc1 signaling cascade. J Immunol, 188(1), 163–169. https://doi.org/10.4049/jimmunol.1101254
[43] Zhou, R. P., Lin, S. J., Wan, W. B., Zuo, H. L., Yao, F. F., Ruan, H. B., et al. (2016). Chlorogenic acid prevents osteoporosis by Shp2/PI3K/Akt pathway in ovariectomized rats. PLoS One, 11(12), e0166751. https://doi.org/10.1371/journal.pone.0166751
[44] Wang, H., Zhao, W., Tian, Q. J., Xin, L., Cui, M., & Li, Y. K. (2020). Effect of lncRNA AK023948 on rats with postmenopausal osteoporosis via PI3K/AKT signaling pathway. Eur Rev Med Pharmacol Sci, 24(5), 2181–2188.
[45] Xi, J. C., Zang, H. Y., Guo, L. X., Xue, H. B., Liu, X. D., Bai, Y. B., et al. (2015). The PI3K/AKT cell signaling pathway is involved in regulation of osteoporosis. J Recept Signal Transduct, 35(6), 640–645. https://doi.org/10.3109/10799893.2015.1041647

This work is licensed under a Creative Commons Attribution 4.0 International License.
Copyright for this article is retained by the author(s), with first publication rights granted to the journal.
This is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).









1.png)














